As a physicist-turned-biologist, I am fascinated by how complex systems function and evolve. My main question is how interactions at one level (e.g., between genes, cells, individuals) create emergent behavior at higher organizational levels (e.g., in genetic networks, multicellular systems, communities). I primarily work on microbial systems and use interdisciplinary methods, combining microfluidics, single-cell imaging, statistical analysis, and mathematical modeling, to create a quantitative understanding of how the properties of microbial communities emerge from intra- and interspecies interactions.

CV
Eduction | ||
| 2017 | Ph.D. in Systems Biology ETH Zurich, Switzerland | |
| 2011 | M.Sc. Cum Laude in Applied Physics Delft University of Technology, Netherlands | |
| 2009 | B.Sc. in Applied Physics Delft University of Technology, Netherlands | |
Research Experience | ||
| 03-2023 present | External Scientific Collaborator University of Lausanne, Switzerland | |
| 01-2022 present | Project Leader | SNSF Ambizione Fellow University of Basel, Switzerland | |
| 05-2020 12-2021 | Postdoc with Urs Jenal University of Basel, Switzerland | |
| 04-2018 03-2020 | Postdoc with Michael Doebeli University of British Columbia, Vancouver, Canada | |
| 08-2017 03-2018 | Postdoc with Martin Ackermann Eawag & ETH Zurich, Switzerland | |
| 02-2013 07-2017 | PhD Student with Martin Ackermann Eawag & ETH Zurich, Switzerland | |
| 09-2012 12-2012 | Visiting Researcher with Peter Galajda Biological Research Centre, Szeged, Hungary | |
| 10-2011 09-2012 | Research Associate with Juan Keymer Delft University of Technology, Netherlands | |
| 05-2011 08-2011 | Intern with Peter Galajda Biological Research Centre, Szeged, Hungary | |
| 03-2010 04-2011 | Master Thesis Student with Juan Keymer and Cees Dekker Delft University of Technology, Netherlands | |
| 02-2008 06-2008 | Bachelor Thesis Student with Teun Klapwijk Delft University of Technology, Netherlands |
PhD and Postdoc Work
Emergent behavior in two species communities
We developed an individual based model to predict how cell-cell interactions affect the dynamics of multispecies microbial communities. We showed that cells exchange metabolites over a short distance of a few cell lengths. This interaction distance can be calculated from a small number of biophysical parameters, and the value predicted by our model closely matched experimental observations.
Read more: Dal Co et al. (2020) | Publisher | pdf
In a follow up project, we developed an analytical framework that quantitively predicts community level properties (equilibrium composition, growth rate, spatial arrangement) from these biophysical parameters. This framework thus allows us to directly link dynamics at the level of molecules to dynamics at the level of the community.
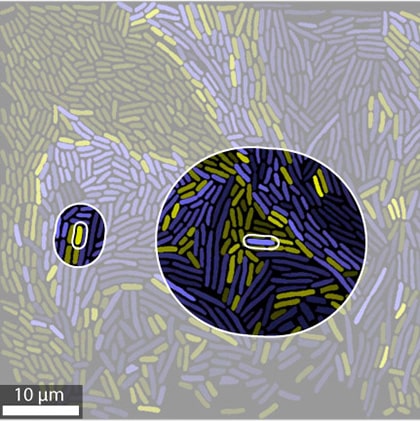
Multilevel selection in microbial communities
We used multilevel selection theory to study the evolutionary dynamics of host associated microbial communities. We showed that microbiomes can evolve to altruistically help their hosts, but only when stringent conditions are met. Specifically, this requires that host have relatively short generation times and strong vertical inheritance of the microbiome.
Read more: van Vliet & Doebeli (2019) | Publisher | pdf
In a follow-up project, we developed an individual based model of free-living microbial communities and showed that multilevel selection can maintain cooperative cross-feeding interactions in multispecies communities. The degree to which cooperators can be maintained depends on the fragmentation mode of the community, and we showed that the fragmentation mode that maximizes the robustness to cheaters is an evolutionary attractor.

Cell-cell interactions in clonal populations
We studied how bacterial colonies organize their activities in space. We found that gene expression levels in bacterial micro-colonies are spatially correlated and developed an analysis method to elucidate the causes of these correlations. We showed that metabolic gradients, history-dependence, and cell-cell interactions were the main factors. Moreover, we found evidence for a novel cell-cell interaction affecting the SOS-response pathway.
Read more: van Vliet et al (2018) | Publisher | pdf
In a second project, we studied how nutrient gradients give rise to metabolic interactions and antibiotic tolerance in bacterial biofilms. We developed a microfluidic device that recreates biofilm like growth conditions within 2D bacterial populations, allowing us to measure growth rates and gene expression levels at the single cell level. We showed that cells create nutrient gradients and as a result they phenotypically differentiate. This leads to metabolic cross-feeding, antibiotic tolerance, and resilience to fluctuating environments.
Read more: Dal Co et al (2019a) | Publisher | pdf
& Dal Co et al (2019b) | Publisher | pdf